Research
Publications
Published in npj Systems Biology and Applications, 2025
Recommended citation: Chevalier, S., Becker, J., Gui, Y. et al. Data-driven inference of Boolean networks from transcriptomes to predict cellular differentiation and reprogramming. npj Syst Biol Appl 11, 105 (2025). https://doi.org/10.1038/s41540-025-00569-z https://doi.org/10.1038/s41540-025-00569-z
Published in CMSB, 2024
Recommended citation: S. Chevalier, D. Boyenval, G. Magaña-López, T. Roncalli, A. Vaginay, L. Paulevé. BoNesis: a Python-based declarative environment for the verification, reprogramming, and synthesis of Most Permissive Boolean networks. CMSB 2024: 22nd International Conference on Computational Methods in Systems Biology, 2024, Pisa, Italy. https://hal.science/hal-04629083
Published in CMSB, 2020
Recommended citation: Chevalier, S., Noël, V., Calzone, L., Zinovyev, A., Paulevé, L. (2020). Synthesis and Simulation of Ensembles of Boolean Networks for Cell Fate Decision. In: Abate, A., Petrov, T., Wolf, V. (eds) Computational Methods in Systems Biology. CMSB 2020. Lecture Notes in Computer Science(), vol 12314. Springer, Cham. doi.org/10.1007/978-3-030-60327-4_11. https://link.springer.com/chapter/10.1007/978-3-030-60327-4_11
Published in ICTAI, 2019
Recommended citation: S. Chevalier, C. Froidevaux, L. Paulevé and A. Zinovyev, "Synthesis of Boolean Networks from Biological Dynamical Constraints using Answer-Set Programming," 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), 2019, pp. 34-41, doi: 10.1109/ICTAI.2019.00014. https://ieeexplore.ieee.org/document/8995442
PhD topic
PhD in computer science for a systems biology issue: modelling of regulatory mechanisms.
I contributed to a method for automatic inference of discrete dynamical models of biological interactions governing complex cell behaviors, called BoNesis. It allows to model regulatory mechanisms of biological processes with complex dynamic properties, such as cell differentiation, by taking into account knowledge on thousands of genes.
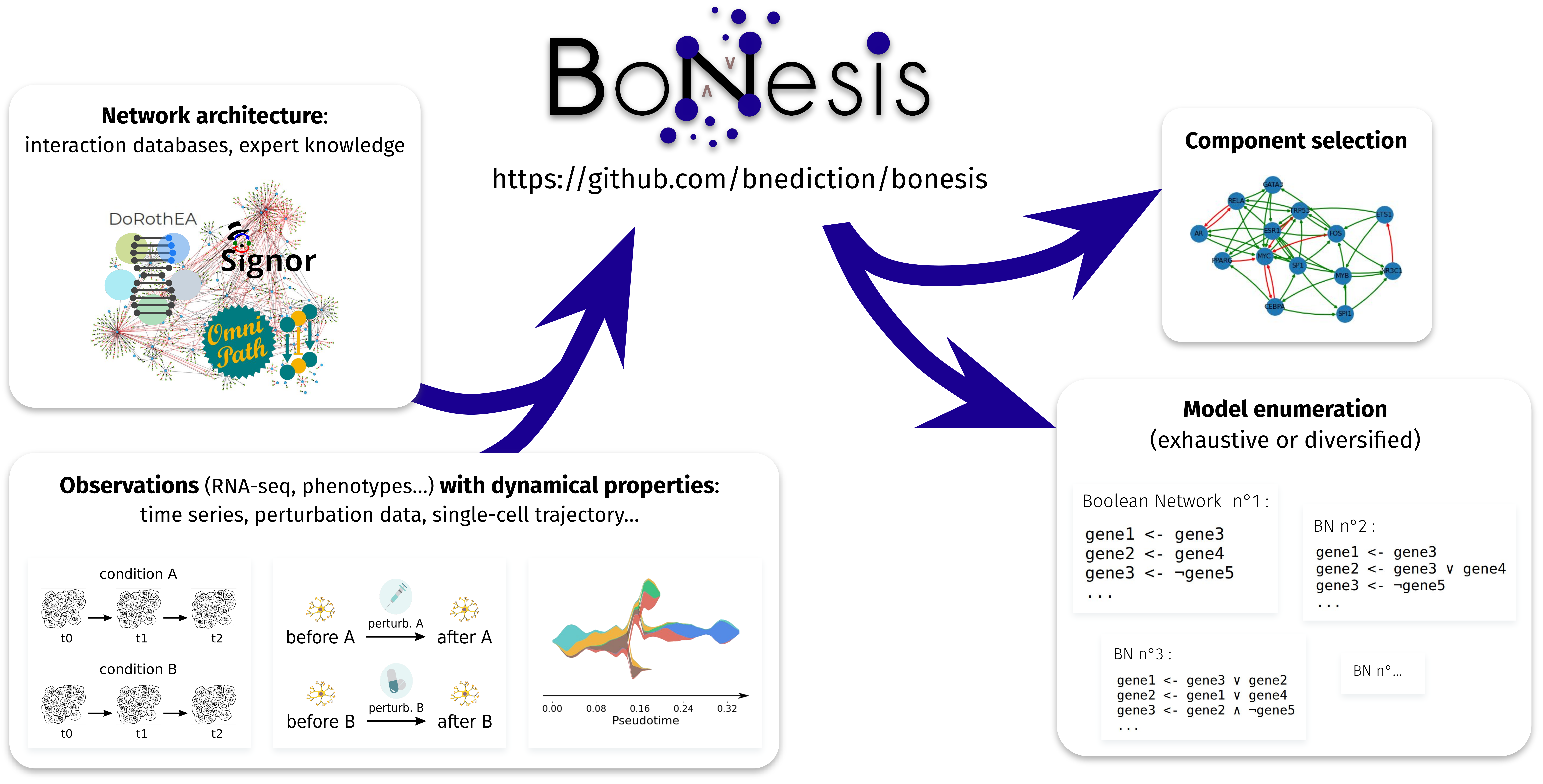
This modeling confronts prior knowledge on interactions with observations along the process to model (bulk/single cell gene expressions, perturbations, mutations…). It can then enumerate all Boolean networks reproducing a complex behavior (under mp semantics), e.g. cell differentiation. It can also be used to help select key regulatory genes among a large gene regulatory network (e.g. from public interaction databases).
Method based on Answer-Set-Programming.
Communications
- Thesis & defense — 09/2022 — Univ. Paris-Saclay
PhD defense - Presentation, international workshop — 07/2021 — Marseille
WAN (Workshop on Automata Networks) - Presentation, national seminar — 03/2021 — virtual
Bioss seminar (CNRS workgroup on systemic and symbolic biology) - Presentation, national workshop — 12/2020 — virtual
SinCellMod-2020 (Single-Cell data in network Modeling) - Presentation, international conference — 09/2020 — virtual
CMSB (int. conf. on Computational Methods in Systems Biology) - Presentation, international research school — 03/2020 — CIRM, Marseille
Network and molecular biology - Presentation, local seminar — 12/2019 — Univ. Paris-Saclay
TheoBioR (CNRS workgroup on metabolism and hybrid methods) - Presentation, international conference — 11/2019 — Portland, Oregon, USA
ICTAI (Int. Conf. on Tools with Artificial Intelligence) - Poster, international conference — 07/2019 — Basel, Swiss
ISMB/ECCB (European Conference on Computational Biology) - Short presentation & poster, international course — 09/2018 — Institut Curie, Paris
CSBC (Computational Systems Biology of Cancer) - Poster, local conference — 09/2018 — Univ. Paris-Saclay
JDSE (Junior Conference on Data Science) - Presentation, national seminar — 07/2018 — Marseille
Bioss annual days (CNRS workgroup on systemic and symbolic biology) - Poster, national conference — 07/2018 — Marseille
JOBIM (Journées Ouvertes en Biologie, Informatique et Mathématiques) </span>

